Front Row L-R: Daniel Garcia, Trung Le, Alp Muyesser, Jonathan Towne, Nikolas Baya, Siddharth Annaldasula, Martha Gahl, Kathleen Callahan, Delaney Moran
Back Row L-R: Lucca Eloy, Chris Ki, Matthew Clapp
Below is the 2017 Summer Undergraduate Research Program in Neuro Computation (uPNC) class roster. Selecting the name of the student will take you to a short description the the student’s research and link to their poster.
- Siddharth Annaldasula
- Nikolas Baya
- Kathleen Callahan
- Matthew Clapp
- Lucca Eloy
- Martha Gahl
- Daniel Garcia
- Chris Ki
- Trung Le
- Delaney Moran
- Alp Muyesser
- Jonathan Towne
The 2017 Summer Schedule can be downloaded here (pdf format).
Siddharth Annaldasula
 Undergraduate Institution: Carnegie Mellon University
Undergraduate Institution: Carnegie Mellon University
Mentor: Andreas Pfenning
University and Department: Carnegie Mellon University, Computational Biology
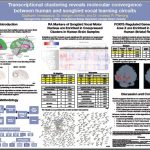
Project Description: Vocal learning is a complex trait evolved in a handful of lineages in mammals and birds, including humans and songbirds. Though phylogenetically distant, the brain circuits for vocal learning in these two groups display remarkable convergence in terms of their anatomy, physiology, and molecular specializations. Previously, the Pfenning research group discovered that multiple human and zebra finch vocal circuit regions that serve analogous functions also display significant convergence in transcriptional correlation, including songbird RA and Area X, and human Laryngeal Motor Cortex and Anterior Striatum, respectively. These transcriptional similarities between analogous brain regions were discovered by comparing the correlation of overall expression between regions. We now improve and expand upon these results by taking a reverse, ‘gene-centric’ approach. We developed computational algorithms to cluster the complete set of 10,783 human genes according to their co-expression across 3,681 human brain regions, and statistically relate the similarity of cluster transcriptional specialization across human and songbird anatomical regions. This set allowed us to identify sets of transcriptional modules that could support the functional properties of analogous human and songbird vocal learning brain regions. These clusters provide insight into the molecular basis for known properties of speech and song-associated brain regions, as well as suggest new molecular pathways that characterize these circuits. These findings provide information on the transcriptome changes that could serve as a causal mechanism in the evolution of vocal learning and other complex behaviors.
Sid performed the following work:
1. Identified groups of genes that are specifically expressed within the neural circuit for vocal production.
2. Benchmarked algorithms that group genes based on their similarity across brain regions.
3. Followed up on candidate genes using experimental methods to measure the expression of individual genes across the brain.
Nikolas Baya
 Undergraduate Institution: Brown University
Undergraduate Institution: Brown University
Mentor: Brent Doiron
University and Department: University of Pittsburgh, Mathematics
Kathleen Callahan
 Undergraduate Institution: University of Texas at Dallas
Undergraduate Institution: University of Texas at Dallas
Mentor: Matthew Smith
University and Department:University of Pittsburgh, Ophthalmology
Project Description:Recent work in systems neuroscience has benefited from molecular tools that identify cell types and allow researchers to tease apart their roles of neural circuits important for sensation, perception and cognition. Extracellular electrophysiology has been unable to take advantage of many of these cellular distinctions, leaving us to only consider neuronal responses on average, and not taking into account separate inhibitory and excitatory neurons, or subtypes of each. Recent work combining electrophysiology and molecular tools in slice physiology has allowed us to identify spiking characteristics of neuronal subtypes, such as waveform shape, firing rate, inter-spike interval, and more. Accessing information about neuronal subtypes in extracellular neurophysiology would enable a host of new investigations into their role in neural circuits that support perception and cognition. Kathleen analyzed large-scale electrophysiological recordings from non-human primate visual and prefrontal cortex, and classified neuronal responses using a multi-dimensional approach. She then used these classifications to determine how different neuronal types play a role in network dynamics and stimulus response in data from animals performing visual tasks.
Matthew Clapp
 Undergraduate Institution: University of South Carolina
Undergraduate Institution: University of South Carolina
Mentor: Tim Verstynen
University and Department: Carnegie Mellon University, Psychology
Project Description: Matthew’s project involved implementing a spiking neural network of the major cortico-basal ganglia (BG) pathways. Matthew investigated the computational role of these pathways by parametrically modulating key parameters of the network — high, medium, and low concentrations of striatal dopamine, for example — and compared the resulting effects on simulated “behavior” generated by the network (e.g., choice accuracy and response time). By incorporating newly discovered knowledge about the architecture of cortico-BG pathways, he addressed a number of outstanding questions regarding the circuit-level interactions between BG nuclei that contribute to adaptive behaviors such as reactive and proactive forms of inhibitory control, decision-making between multiple alternatives, and feedback-dependent learning. Matthew took a lead role in programming the neural network and assisted in building a framework for manipulating network parameters, simulating virtual experiments, and analyzing and visualizing network-generated behavioral data.
Lucca Eloy
 Undergraduate Institution: College of the Holy Cross
Undergraduate Institution: College of the Holy Cross
Mentor: Rob Kass
University and Department: Carnegie Mellon University, Statistics
Project Description:Lucca examined trial-to-trial variation in Evoked-Related Potentials (ERPs) in magnetoencephalography (MEG). In MEG, source localization is used to infer signals originating across the brain from external sensor recordings. An advantage of MEG is its millisecond temporal resolution. Many studies examine “functional connectivity” across brain regions using a pair of techniques, Granger causality and phase locking, which turn out to be susceptible to distortion due to ERPs. Two other members of the Kass research group have been investigating this issue in local field potentials (LFPs), and their methods should apply also to MEG. However, MEG source signals are noisier than LFPs. Preliminary work is needed to understand the degree of the trial-to-trial variation. This is the work Lucca did: Basic descriptive methods, i.e. plots and elementary data summaries. Lucca also applied tools such as regression analysis and more.
Martha Gahl
 Undergraduate Institution: University of Missouri
Undergraduate Institution: University of Missouri
Mentor: Stephen Meriney
University and Department: University of Pittsburgh, Neuroscience
Project Description: Martha modeled the calcium-triggered transmitter release from the motor nerve terminal using the microphysiological simulation environment MCell. This is a particle-based diffusion-reaction simulator. Martha tested the role of changing the number and distribution of presynaptic calcium channels in transmitter output from these synapses. Our research group will use these simulation data in comparison with physiology recordings from live neuromuscular synapses to constrain future models and experiments. This approach will allow us to predict sub-cellular events that govern the function of synapses. Martha learned how to use the MCell model environment, made changes to the input files that govern each modeling run, learned how to analyze data output from the model, and tested several hypotheses regarding the organization of transmitter release sites and their role in synaptic function.
Daniel Garcia
 Undergraduate Institution: University of Pittsburgh
Undergraduate Institution: University of Pittsburgh
Mentor: Tracy Cui
University and Department: University of Pittsburgh, Bioengineering
Chris Ki
 Undergraduate Institution: University of California at Berkeley
Undergraduate Institution: University of California at Berkeley
Mentor: Aryn Gittis
University and Department: Carnegie Mellon University, Biological Sciences
Trung Le
 Undergraduate Institution: Texas A&M University
Undergraduate Institution: Texas A&M University
Mentor: Steve Chase
University and Department: Carnegie Mellon University, Biomedical Engineering
Project Description: The Chase research group uses brain-computer interfaces (BCIs) to study motor learning. Trung developed computational techniques for assessing short timescale correlations between neurons recorded over multiple days while the subject engaged in a BCI learning task. He then compared the functional connectivity of neurons directly involved in BCI control to neurons that were not engaged in direct control.
Delaney Moran
 Undergraduate Institution: Middlebury College
Undergraduate Institution: Middlebury College
Mentor: Jennifer Collinger
University and Department: University of Pittsburgh, Physical Medicine and Rehabilitation
Project Description: Delaney collected data as part of an ongoing human brain-computer interface (BCI) trial where signals are recorded from motor cortex of a human tetraplegic participant and sensory feedback is provided by microstimulation through electrodes implanted the participant’s somatosensory cortex. A particular focus of her project was improving the participant’s ability to grasp objects. Delaney quantified changes in neural activity that was recorded while the participant was grasping an object as compared to making the same movement without an object in the workspace. Additionally, Delaney collected grasp force data from able-bodied participants to determine how performance is influenced by task instructions and design. Delaney tracked signal quality from an implanted electrode, as this is known to change over the lifetime of the implant.
Alp Muyesser
 Undergraduate Institution: Carnegie Mellon University
Undergraduate Institution: Carnegie Mellon University
Mentor: Tim Verstynen
University and Department: Carnegie Mellon University, Psychology
Jonathan Towne
 Undergraduate Institution: Rice University
Undergraduate Institution: Rice University
Mentor: Avniel Ghuman
University and Department: University of Pittsburgh, Neurological Surgery

